At VTT we are able to identify novel candidate enzymes to help metabolic engineering by conducting homology based database searches. Public and in-house databases are queried with sequences reported to have the interesting activity.
The homology based searches are conducted against Uniprot (SwissProt and TrEMBL) and GenBank protein databases (nr, pat and env_nr) using blastp, and against GenBank nucleotide databases (tsa_nt, env_nt and pat) and our in-house nucleotide databases using tblastn. The GenBank pat databases contain sequences from patents, deposited by the U.S. Patent and Trademark Office (Benson et al., 2013). GenBank tsa_nt and env_nt databases contain DNA sequences assembled based on shotgun sequencing data, such as RNA sequencing or metagenomics sequencing (Benson et al., 2013). Sequences with E-values smaller or equal to 1e-30 are extracted in each case.
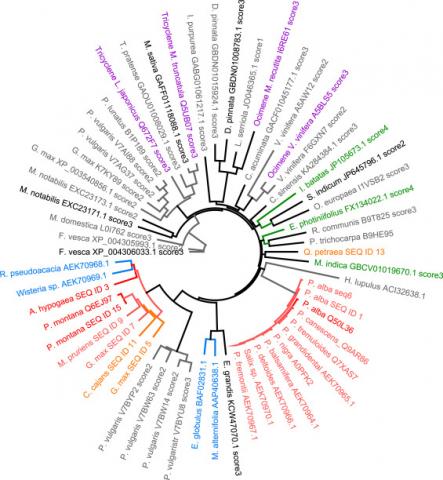
Multiple sequence alignment and phylogenetic tree constructionThe retrieved sequences are analysed based on a multiple sequence alignment (MSA) and a phylogenetic tree. The MSA is created by aligning the protein sequences to the protein family Pfam motif using the HMMer programme (Eddy, 1998). The MSA is used as an input for the phylogenetic tree reconstruction algorithm FastTree.
Solutions offered at VTT
As a result of our analysis we provide to our customers a phylogenetic tree of potential candidate sequences together with the reference enzymes where the protected sequences are identified.
Our method for novel enzyme search has been successfully applied in many customer project. The method is described in:
Identification of novel isoprene synthases through genome mining and expression in Escherichia coli